Technical Documentation
Introduction
MDSE is a high-performance computational framework designed to orchestrate parallel molecular dynamics (MD) simulations. It automates the interaction with databases, manages complex simulation lifecycles, and computes material properties for high-throughput analysis.
By integrating robust open-source libraries, MDSE provides a streamlined interface for simulating crystal structures and analyzing defects.
Architecture & Tech Stack
MDSE is built upon a modular architecture, leveraging specific libraries for simulation, parallelization, and data management.
Simulation Engine
ASE (Atomic Simulation Environment)
The core of the simulation framework [1]. MDSE utilizes the ASE Atoms object as the primary data structure for atomic positions, properties, and metadata. The ASE Calculator interface allows MDSE to seamlessly integrate various interatomic potentials.
ASAP (As Soon As Possible) Used as a high-performance extension to ASE [2]. While ASE handles the structure, ASAP is employed for efficient force evaluations and optimized potential performance. It is particularly critical for large-scale systems where standard ASE calculators may be the bottleneck.
Data Management
HTTk (High-Throughput Toolkit)
MDSE utilizes the httk.db module to interface with SQLite databases containing material defect data [3]. This allows for the automated retrieval and setup of simulation environments based on existing material datasets.
PyMongo The bridge between MDSE and the MongoDB storage backend. It handles the serialization of simulation results and material properties into the document-based database.
Optimade API Ensures that all data outputs comply with the FAIR (Findable, Accessible, Interoperable, and Reusable) data principles, ensuring compatibility with the broader materials science ecosystem.
Parallelization
MPI (Message Passing Interface) To maximize computational efficiency, MDSE implements parallelization via MPI. This allows the software to distribute multiple simulations across multiple CPU cores, significantly reducing runtime for large simulation batches.
System Overview
MDSE is architected into two primary, decoupled modules. This separation of concerns improves maintainability and allows for independent scaling of simulation and analysis capabilities.
Simulation Module: The execution engine. It accepts user configurations, initializes the ASE/ASAP environment, and manages the MD lifecycle.
Data Processing Module: The analytical engine. It handles database I/O, post-processing of trajectories, and visualization.
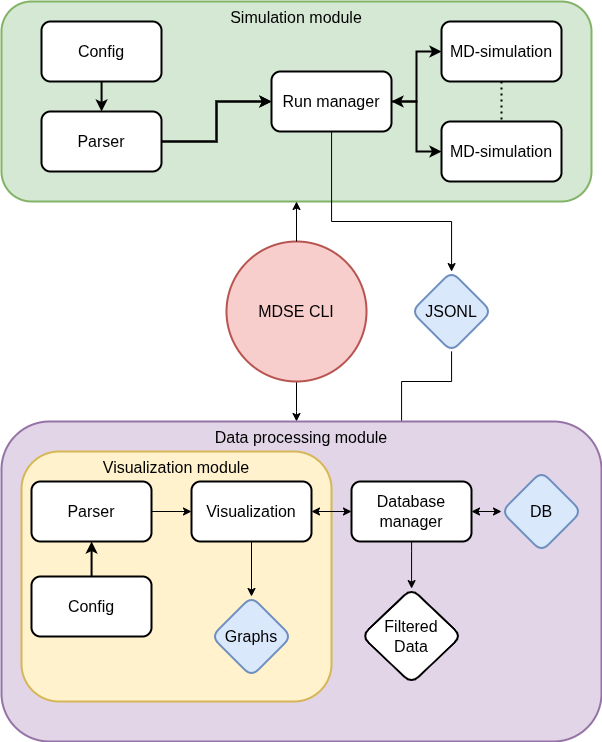
Simulation Workflow
The MDSE workflow is designed to be linear and reproducible. The process follows a clear path from Configuration -> Initialization -> Execution -> Analysis.
1. Configuration (CLI)
MDSE operates primarily via a Command Line Interface (CLI). Users initiate simulations by passing a configuration file (YAML format) to the mdse.parser.
This configuration defines:
Crystal Properties: Structure files, supercell size, etc.
Simulation Parameters: Temperature, time steps, ensemble (NVT/NVE…), etc.
Output Requests: Specific material properties to calculate.
Note: CLI flags are available to override specific parameters in the config file without editing the source YAML.
2. Initialization & Parsing
The Parser module validates the input and forwards the data to the RunManager.
The Run Manager instantiates SimulationManager objects. These objects encapsulate the simulation state, attaching the appropriate potentials (Calculators) to the ASE Atoms objects.
3. Execution
If the input source is an external database (e.g., ADAQ [4]), MDSE queries the SQLite database for structures matching the criteria. These are extracted to .cif files.
The simulation is then executed using the combined power of ASE and ASAP, utilizing MPI for parallel processing where applicable.
4. Calculation & Analysis
Upon simulation completion, the Run Manager invokes ResultMD objects.
Equilibrium Check: Data is first filtered to remove non-equilibrated frames (burn-in period) to prevent skewed results.
Property Extraction: Physical properties are calculated as time-averages over the remaining valid trajectory frames.
5. Storage
Results are committed to the storage backend.
MongoDB: Hosted locally (via Docker), this serves as the primary sink for processed simulation results.